Lungs and Lobes Semantic Segmentation in Mediastinal CT Scans Using 3D Convolutional Neural Networks
Master's thesis by Kristin Schive Hjelde
Abstract
Pulmonary CT image analysis is a vital part in the assessment and treatment planning of different lung diseases. The method often requires the lungs to be separated from the surrounding structures, a process known as lung segmentation. Fissures divide the lungs into smaller compartments known as lobes, with their own independent vessels and airways system. Some diseases develop only in one of these lobes, and some treatments are performed on a lobar level. It is thus necessary to also segment the lobes in many cases. Doing lungs and lobes segmentation by hand is a tedious and time-consuming process, requiring expert radiologists. A variety of different methods to automate the segmentation process have been proposed throughout the years, but many of them struggle on lungs with high amounts of abnormalities, which often is the case for diseased lungs.
This thesis aims at contributing further towards the full automation of lung and lobe segmentation by investigating two state-of-the-art neural network architectures, and their ability to generate accurate segmentation models with short training and inference time. The two studied approaches are the 3D U-Net, and the relatively new PLS-Net. Both are 3D fully convolutional networks whereby the U-Net is more traditional with a large number of parameters while the PLS-Net is extremely lightweight in comparison. The two networks were tested for the lung segmentation task, using different combinations of deep learning frameworks (e.g., Tensorflow and PyTorch), training precision, batch sizes and input resolutions. In addition, the PLS-Net was trained and tested for the lobe segmentation task, together with a simple post-processing step.
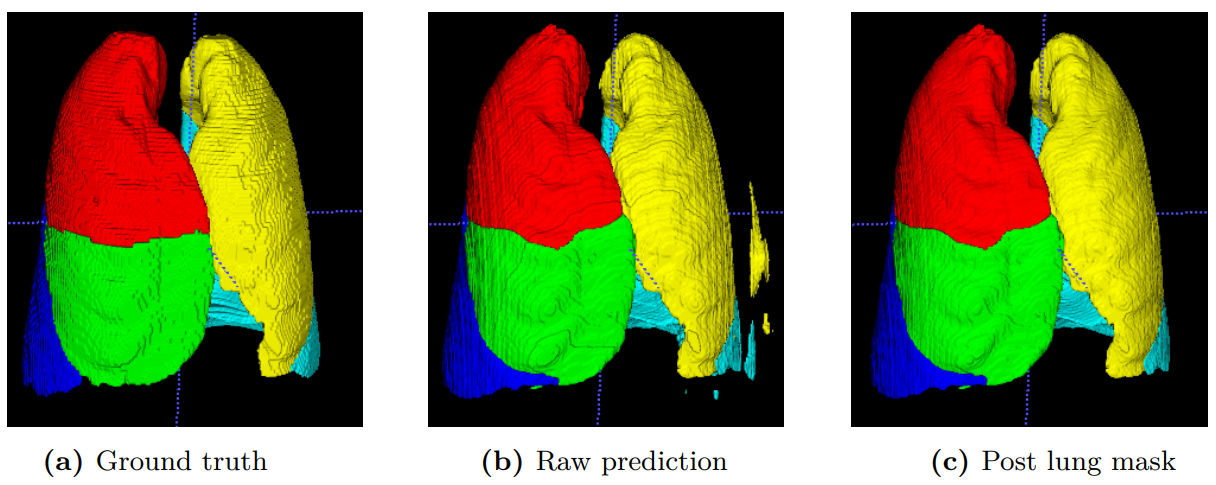
The results showed that using smaller input volumes with batch size 2 gave higher accuracy than using larger input volumes with batch size 1. PyTorch and TensorFlow gave equally good Dice scores on the lung segmentation task, and PyTorch gave better training performance, but was slower during inference. Using mixed precision over full precision reduced the memory footprint by 40%, without reducing the accuracy. The PLS-Net used 30% less memory than the U-Net for the same input data and batch size, with a reduction of 0.3% in Dice score. For the lobe segmentation task, the PLS-Net gave a Dice score of 92.6% before post-processing, and 92.9% after. The lobe segmentation model performed equally well on data from another data set and the data used for training, but struggled on samples containing abnormalities.
Supervised by David Bouget, André Pedersen, and Ilangko Balasingham